ChIP analysis of histone modifications at the Ig e locus in CL-01 cells
Par un écrivain mystérieux
Description
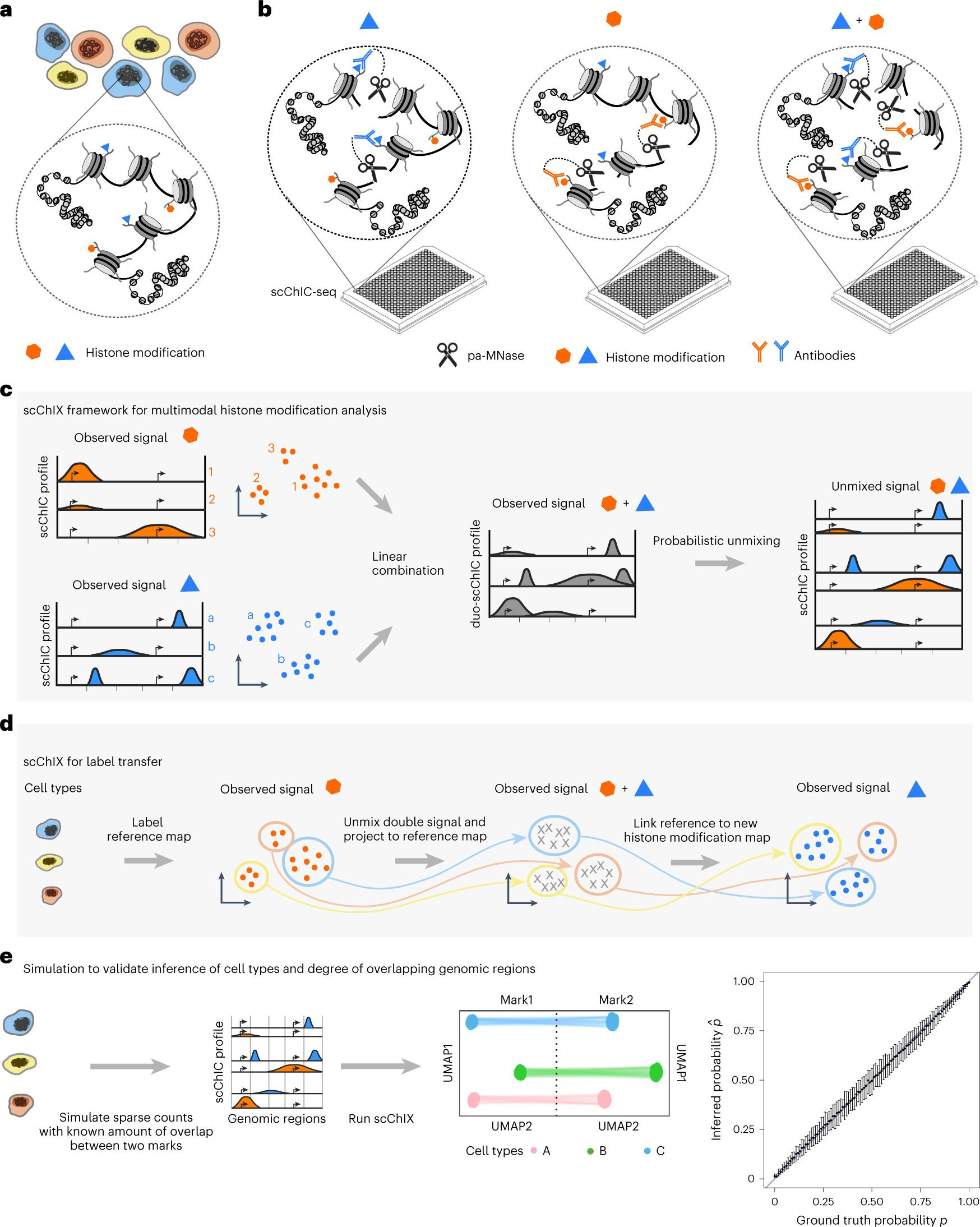
scChIX-seq infers dynamic relationships between histone modifications in single cells

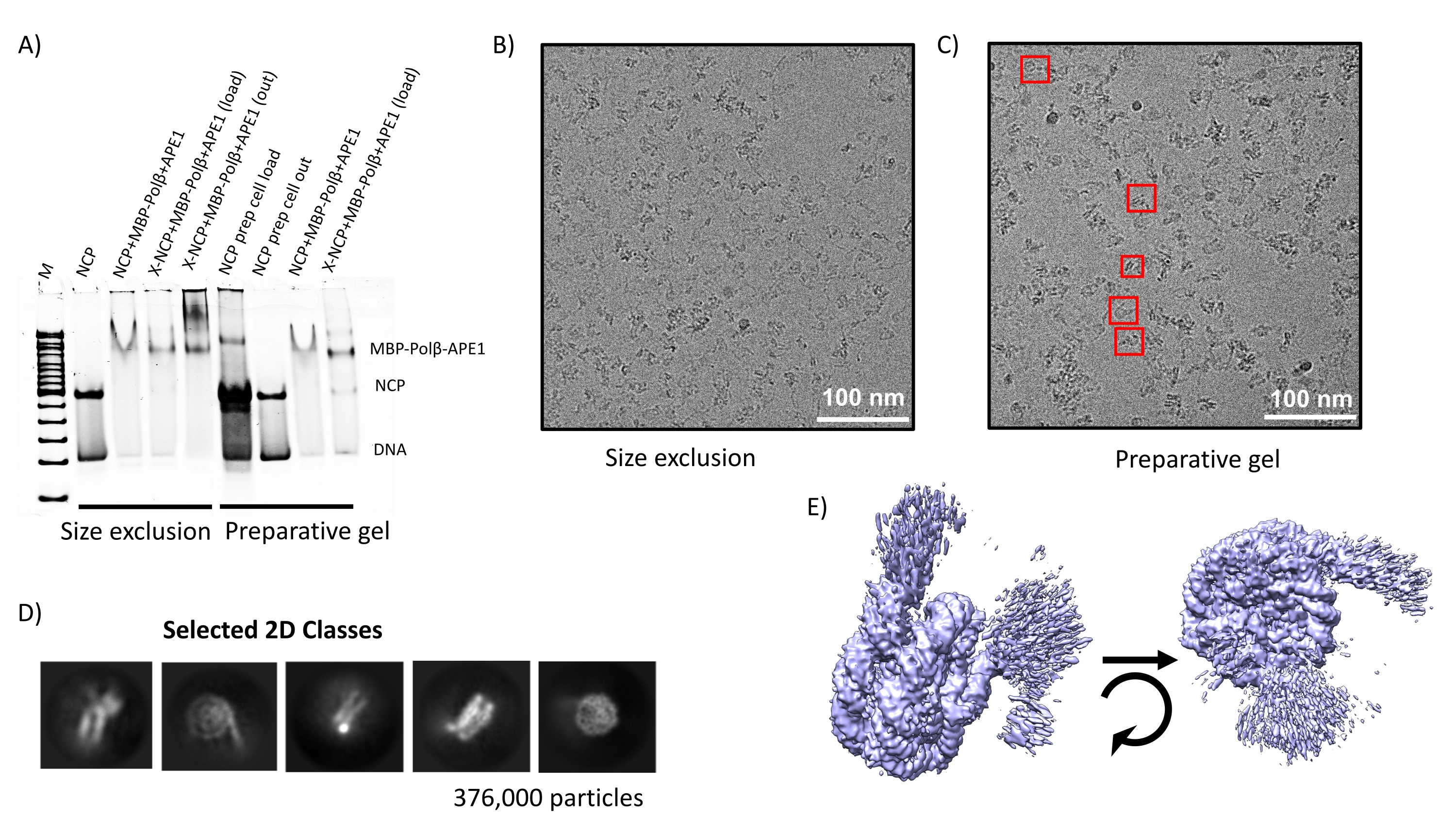
Phf5a regulates DNA repair in class switch recombination via p400 and histone H2A variant deposition

Novel cell adhesion/migration pathways are predictive markers of HDAC inhibitor resistance in cutaneous T cell lymphoma - eBioMedicine

Lactate induces metabolic and epigenetic reprogramming of pro‐inflammatory Th17 cells

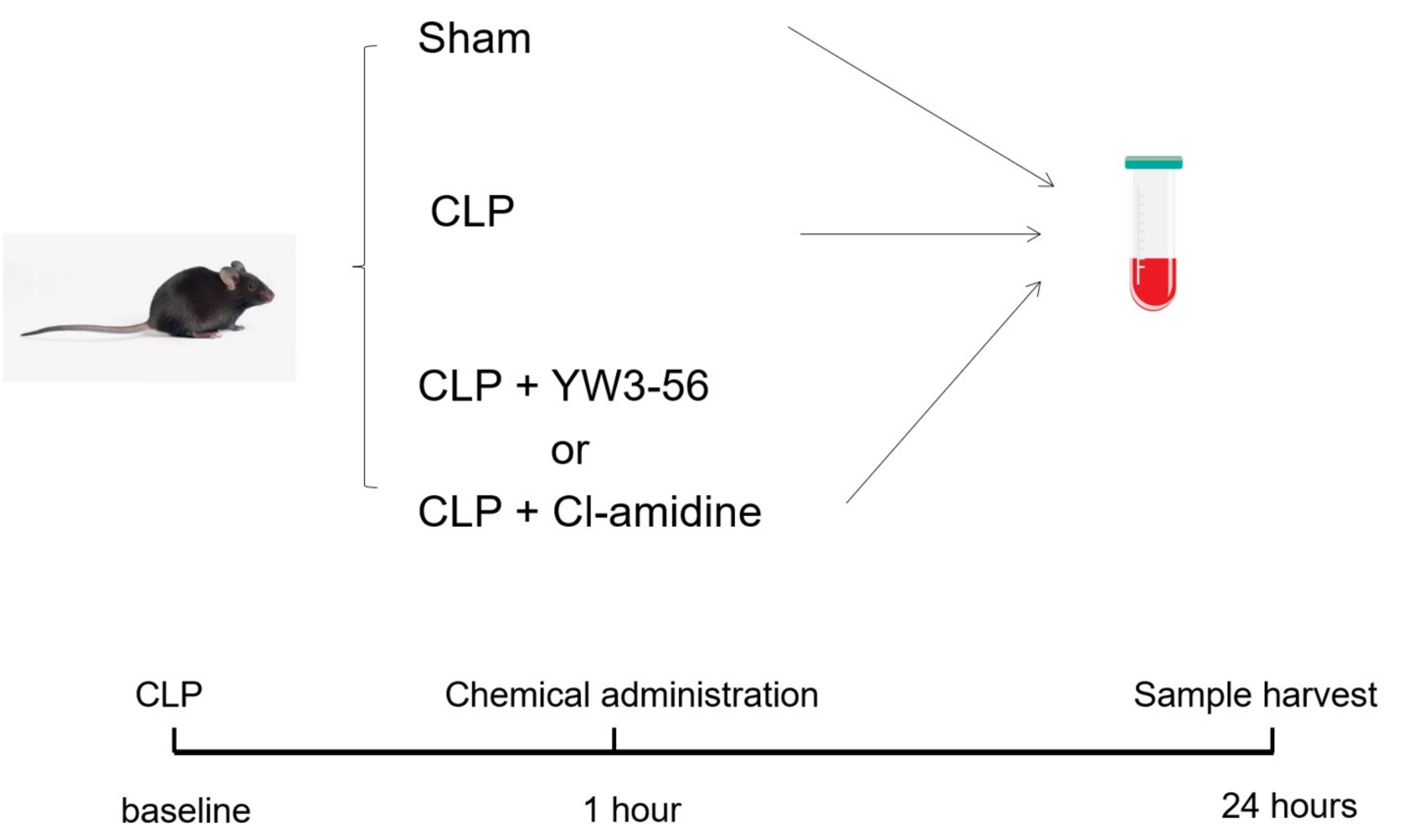
IJMS, Free Full-Text

scChIX-seq infers dynamic relationships between histone modifications in single cells

Epigenomic analysis reveals DNA motifs regulating histone modifications in human and mouse

Cancers, Free Full-Text

Epigenomic mapping reveals distinct B cell acute lymphoblastic leukemia chromatin architectures and regulators - ScienceDirect

Multiscale 3D genome reorganization during skeletal muscle stem cell lineage progression and aging
depuis
par adulte (le prix varie selon la taille du groupe)